-
Viewpoint on 'IONS'
Viewpoint on 'Scientific Literacy'
- Proudly sponsored by
-

-
Subnanometer Perfection

According to French philosopher Voltaire (1694-1778) the perfect is the enemy of the good, thus not exactly desirable. As recent results show, however, where scientific research is concerned this may not always hold true.
-
Tiny Mirrors in the Sky to Fight Global Warming

Tiny particles in the atmosphere play a decisive role in determining the Earth’s temperature. Can nanoparticles be deployed to fight global warming?
-
Solar Fuel: No More Drilling!

The Earth’s reserve of fossil fuels is limited, and their production and use pollute our environment. Solar collectors offer a possible solution to the increasingly pressing demands for economically and environmentally sustainable energy.
Volume 9 Story 7 - 30/6/2010
"I am afraid you are going to need an x-ray." This is probably the last thing you want to hear from your physician after a bad fall. X-rays can help us see inside solid objects and, therefore, the tool of election in medical diagnostic radiography. Less known is, perhaps, the fact that x-rays can be used for high-resolution imaging of biological samples. Images of whole yeast cells, with the stunning resolution of ten nanometers, have now been acquired by employing lensless x-ray diffraction microscopy: this is the highest resolution ever obtained with this method for biological samples.
This work was directed by Chris Jacobsen at the Northwestern University (Evanston, Illinois) and was carried out by researchers at Stony Brook University (Stony Brook, New York), in a joint effort with researchers at Lawrence Berkeley National Laboratory (Berkeley, California), and at Argonne National Laboratory (Argonne, Illinois).
Since its invention by Dutchman Antonie van Leeuwenhoek and Englishman Robert Hooke more than three hundred years ago, the optical microscope has been the standard tool for cell imaging. Unfortunately, the information it can deliver is limited by a fundamental physical law: the limit of diffraction. According to this law, it is not possible to distinguish two objects separated by less than half the wavelength of the illumination, which is a few hundred nanometers in the case of light. More refined instruments are needed to break the diffraction limit barrier.
X-rays are a form of electromagnetic radiation, as are microwaves or visible light. However, while visible light has a wavelength in the order of a few hundred nanometers, the wavelength of x-rays is much shorter, ranging from between the size of an average molecule (10 nanometers) to less than a fraction of an atom (0.01 nanometers). X-rays were discovered by German physicist Wilhelm Conrand Röntgen in 1895 - a discovery which earned him the very first Nobel Prize in Physics in 1901. Common classification separates between hard x-rays, with shorter wavelengths and higher energy, and soft x-rays, with longer wavelengths and lower energy.
An established technique takes advantage of the high resolution and deep penetration ability of x-rays to image in 3D cells floating in water, and therefore under conditions close to their natural ones: a large number of cells can be imaged in a short time at resolutions of 40 to 60 nanometers. However, pushing down this resolution is limited by x-ray optics. "There are a number of different types of optics used with x-rays: zone plates, K-B mirrors, capillaries, compound refractive lenses, to name but a few," points out Johanna Nelson at Stony Brook University [1]. "Each has its own set of pros and cons, but none of them are the silver bullet to x-rays which optical lenses are to visible light." Even the best x-ray lenses are not very efficient: higher resolution images would therefore need a very intense radiation exposure, thus damaging the biological specimen too quickly.
The technique developed by the researchers led by Jacobsen works without lenses, as it replaces its function with a mathematical algorithm. "The x-rays leaving the sample that we would like to image are described as waves with both a magnitude and phase," Nelson explains. "Typically a lens converts this information into a real space image. Mathematically the job of the lens can be replaced by a single Fourier transform. However, the CCD detector which measures the scattered x-rays only measures their intensity — magnitude squared — so the phase information is lost. Without this phase information, the single Fourier transform will not produce an image. The reconstruction process is an iterative algorithm to recover that lost phase." By doing so, it has been possible to achieve high resolution images – of down to ten nanometers – something impossible for lens-based techniques. These results also suggest that by employing the intrinsic x-ray ability of penetration, a full 3D cell tomography should soon be obtainable, thus helping reveal valuable information about the relationship of a cell’s internal structure.
X-ray imaging of biological specimens competes with other high-resolution techniques. For example, electron beam microscopy has been widely employed in biology to gain resolution at the nanoscale. Indeed, the wavelength associated with fast electrons is much smaller than that of light — even smaller than that of x-rays. These unconventional microscopes can easily achieve a resolution of ten nanometers or less, but they are limited to very thin samples, of a few hundred nanometers or less; they cannot, consequently, be used to look through a whole cell.
Another way of beating the limit of diffraction of light is by taking advantage of fluorescence, an optical property of some molecules. Fluorescent molecules absorb light at a certain wavelength to emit it again at a longer one. High spatial resolution down to a few tens of nanometers has been achieved through several fluorescence-based techniques, such as STED or STORM [2-4]. "Both STED and STORM require the use of fluorescence tags," Nelson says. "These tags are useful in identifying the location of specific molecules; however, multiple tags must be used to put these molecules in the larger cellular context. X-ray imaging uses a smaller wavelength, and easily achieves sub-100 nanometer resolution and the energy can be tuned to pick out the natural contrast between materials. Light microscopy will always have the advantage of live cell imaging, which is impossible with the dose required for high-resolution x-ray imaging. However, frozen hydrated imaging with x-rays is a nice alternative which allows the cells to be frozen in their natural hydrated state, while the sample is much more resistant to radiation. Furthermore, labels are not required for reconstruction. In fact, previous publications demonstrating this technique on yeast cells did not use labels [5-6]."
The new high-resolution x-ray diffraction microscopy can certainly help shed light on the cell’s internal structure. "This technique could be used in 3D as an alternative to lens-based techniques for samples where radiation dose is a concern," Nelson envisions. "It is particularly relevant for imaging thick biological samples which would have to be sliced very thin to be imaged in an electron microscope. This is why we chose to image a eukaryotic cell and not something smaller, such as a bacterium or a virion."
[1] J.N. wishes to acknowledge and give due credit to the work of the coauthors of the paper, and in particular to the work of Xiaojing Huang and Jan Steinbrener.
[2] S. Hell, Far-Field Optical Nanoscopy, Science 316, 1153-1158 (2007).
[3] N. van Hulst, Many Photons get More out of Diffraction, Optics & Photonics Focus 4, 1 (2009) (link).
[4] G. Volpe, Nanoscopy: Shedding Light on Life, Optics & Photonics Focus 1, 4 (2008) (link).
[5] D. Shapiro, P. Thibault, T. Beetz, V. Elser, M. Howells, C. Jacobsen, J. Kirz, E. Lima, H. Miao, A. M. Neiman, and D. Sayre, Biological Imaging by Soft X-Ray Diffraction Microscopy, PNAS 102, 15343-15346 (2005).
[6] X. Huang, J. Nelson, J. Kirz, E. Lima, S. Marchesini, H. Miao, A. M. Neiman, D. Shapiro, J. Steinbrener, A. Stewart, J. J. Turner, and C. Jacobsen, Soft X-Ray Diffraction Microscopy of a Frozen Hydrated Yeast Cell, Physical Review Letters 103, 198101 (2009).

Cells through X-Rays
Thanks to their ability to see through solid objects, X-rays are one of the most powerful tools of modern medicine. They may soon enable us to see inside a single cell with a stunning resolution of ten nanometers.
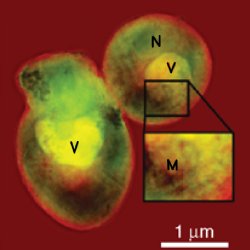
An x-ray view on a yeast cell. By employing lensless x-ray diffraction microscopy, this image of a group of yeast cells has been acquired with a resolution of ten nanometers — approximately the length of 50 water molecules in a row. This is the highest resolution ever obtained with this method for biological samples. In this image the letters represent possible structures of the cell, such as vacuoles (V), mitochondria (M), and nucleus (N). Courtesy of Johanna Nelson.
This work was directed by Chris Jacobsen at the Northwestern University (Evanston, Illinois) and was carried out by researchers at Stony Brook University (Stony Brook, New York), in a joint effort with researchers at Lawrence Berkeley National Laboratory (Berkeley, California), and at Argonne National Laboratory (Argonne, Illinois).
Since its invention by Dutchman Antonie van Leeuwenhoek and Englishman Robert Hooke more than three hundred years ago, the optical microscope has been the standard tool for cell imaging. Unfortunately, the information it can deliver is limited by a fundamental physical law: the limit of diffraction. According to this law, it is not possible to distinguish two objects separated by less than half the wavelength of the illumination, which is a few hundred nanometers in the case of light. More refined instruments are needed to break the diffraction limit barrier.
X-rays are a form of electromagnetic radiation, as are microwaves or visible light. However, while visible light has a wavelength in the order of a few hundred nanometers, the wavelength of x-rays is much shorter, ranging from between the size of an average molecule (10 nanometers) to less than a fraction of an atom (0.01 nanometers). X-rays were discovered by German physicist Wilhelm Conrand Röntgen in 1895 - a discovery which earned him the very first Nobel Prize in Physics in 1901. Common classification separates between hard x-rays, with shorter wavelengths and higher energy, and soft x-rays, with longer wavelengths and lower energy.
An established technique takes advantage of the high resolution and deep penetration ability of x-rays to image in 3D cells floating in water, and therefore under conditions close to their natural ones: a large number of cells can be imaged in a short time at resolutions of 40 to 60 nanometers. However, pushing down this resolution is limited by x-ray optics. "There are a number of different types of optics used with x-rays: zone plates, K-B mirrors, capillaries, compound refractive lenses, to name but a few," points out Johanna Nelson at Stony Brook University [1]. "Each has its own set of pros and cons, but none of them are the silver bullet to x-rays which optical lenses are to visible light." Even the best x-ray lenses are not very efficient: higher resolution images would therefore need a very intense radiation exposure, thus damaging the biological specimen too quickly.
The technique developed by the researchers led by Jacobsen works without lenses, as it replaces its function with a mathematical algorithm. "The x-rays leaving the sample that we would like to image are described as waves with both a magnitude and phase," Nelson explains. "Typically a lens converts this information into a real space image. Mathematically the job of the lens can be replaced by a single Fourier transform. However, the CCD detector which measures the scattered x-rays only measures their intensity — magnitude squared — so the phase information is lost. Without this phase information, the single Fourier transform will not produce an image. The reconstruction process is an iterative algorithm to recover that lost phase." By doing so, it has been possible to achieve high resolution images – of down to ten nanometers – something impossible for lens-based techniques. These results also suggest that by employing the intrinsic x-ray ability of penetration, a full 3D cell tomography should soon be obtainable, thus helping reveal valuable information about the relationship of a cell’s internal structure.
X-ray imaging of biological specimens competes with other high-resolution techniques. For example, electron beam microscopy has been widely employed in biology to gain resolution at the nanoscale. Indeed, the wavelength associated with fast electrons is much smaller than that of light — even smaller than that of x-rays. These unconventional microscopes can easily achieve a resolution of ten nanometers or less, but they are limited to very thin samples, of a few hundred nanometers or less; they cannot, consequently, be used to look through a whole cell.
Another way of beating the limit of diffraction of light is by taking advantage of fluorescence, an optical property of some molecules. Fluorescent molecules absorb light at a certain wavelength to emit it again at a longer one. High spatial resolution down to a few tens of nanometers has been achieved through several fluorescence-based techniques, such as STED or STORM [2-4]. "Both STED and STORM require the use of fluorescence tags," Nelson says. "These tags are useful in identifying the location of specific molecules; however, multiple tags must be used to put these molecules in the larger cellular context. X-ray imaging uses a smaller wavelength, and easily achieves sub-100 nanometer resolution and the energy can be tuned to pick out the natural contrast between materials. Light microscopy will always have the advantage of live cell imaging, which is impossible with the dose required for high-resolution x-ray imaging. However, frozen hydrated imaging with x-rays is a nice alternative which allows the cells to be frozen in their natural hydrated state, while the sample is much more resistant to radiation. Furthermore, labels are not required for reconstruction. In fact, previous publications demonstrating this technique on yeast cells did not use labels [5-6]."
The new high-resolution x-ray diffraction microscopy can certainly help shed light on the cell’s internal structure. "This technique could be used in 3D as an alternative to lens-based techniques for samples where radiation dose is a concern," Nelson envisions. "It is particularly relevant for imaging thick biological samples which would have to be sliced very thin to be imaged in an electron microscope. This is why we chose to image a eukaryotic cell and not something smaller, such as a bacterium or a virion."
[1] J.N. wishes to acknowledge and give due credit to the work of the coauthors of the paper, and in particular to the work of Xiaojing Huang and Jan Steinbrener.
[2] S. Hell, Far-Field Optical Nanoscopy, Science 316, 1153-1158 (2007).
[3] N. van Hulst, Many Photons get More out of Diffraction, Optics & Photonics Focus 4, 1 (2009) (link).
[4] G. Volpe, Nanoscopy: Shedding Light on Life, Optics & Photonics Focus 1, 4 (2008) (link).
[5] D. Shapiro, P. Thibault, T. Beetz, V. Elser, M. Howells, C. Jacobsen, J. Kirz, E. Lima, H. Miao, A. M. Neiman, and D. Sayre, Biological Imaging by Soft X-Ray Diffraction Microscopy, PNAS 102, 15343-15346 (2005).
[6] X. Huang, J. Nelson, J. Kirz, E. Lima, S. Marchesini, H. Miao, A. M. Neiman, D. Shapiro, J. Steinbrener, A. Stewart, J. J. Turner, and C. Jacobsen, Soft X-Ray Diffraction Microscopy of a Frozen Hydrated Yeast Cell, Physical Review Letters 103, 198101 (2009).
Giorgio Volpe
2010 © Optics & Photonics Focus
GV is currently working on his doctoral thesis at ICFO - The Institute of Photonic Sciences, Barcelona (Spain).

Johanna Nelson, Xiaojing Huang, Jan Steinbrener, David Shapiro, Janos Kirz, Stefano Marchesini, Aaron M. Neiman, Joshua J. Turner & Chris Jacobsen, High-resolution x-ray diffraction microscopy of specifically labeled yeast cells, PNAS (2010) 107, 235-7239 (link).